|
The TEDDY Study is committed to maximizing its contribution to the greater scientific community. Beyond published findings, the TEDDY Study may also offer researchers everything from methods and technologies developed on study to the actual data and samples collected from participants.
Data Assets
A variety of different data types are collected as a part of the TEDDY study, including clinical metadata and laboratory test result data across various 'omics analytes. The TEDDY Data Coordinating Center (DCC) manages, curates, integrates, and provisions these data assets for analysis by TEDDY and approved external investigators. Existing TEDDY clinical and ‘omics data assets are summarized below and detailed in the linked documents.
Clinical Metadata: |
'Omics Analytes: |
Documentation: |
- Antibodies
- Demographics
- Diet
- Genotypes
- Case-Control Indicators
- Household Exposures
- Medications
- Medical History
- Family History
- Pre and Perinatal Exposures
- Psychosocial Stressors
|
- Dietary Biomarkers
- Exome
- Gene Expression
- Lipidomics
- Metabolomics
- Microbiome and Metagenomics
- Proteomics
- RNA Sequencing
- SNPs
- Whole Genome Sequencing
|
|
Data Sharing
The TEDDY Study has adopted policies and procedures in support of its commitment to sharing data with the scientific community while also protecting the privacy of participants. In accordance with the TEDDY Data Sharing Policy, data are made publicly available via designated controlled-access data repositories. A detailed listing of publicly available TEDDY data assets and instructions for accessing them via the relevant data repositories can be found in the links below. These data releases have been submitted at different time points and to various repositories, depending on NIH requirements and the nature of the data. Each submission is treated as an independent release, possessing uniquely masked subject and sample identifiers. Researchers may desire to combine data across these releases for analysis, but are unable to do so as a result of the independently masked identifiers. The NIDDK repository can now provide repository data release identifier mapping materials to satisfy this demand once the investigators have received approval to access the data. In addition, external investigators can obtain access to TEDDY 'omics data and the TEDDY Web Portal for analysis (see Data Infrastructure below) via application to the TEDDY Ancillary Studies Committee.
TEDDY samples can also be made available to researchers as part of an ancillary study following the submission and approval of a proposal to the TEDDY Ancillary Studies Committee. Please email TEDDY@epi.usf.edu for more details.
Data Infrastructure
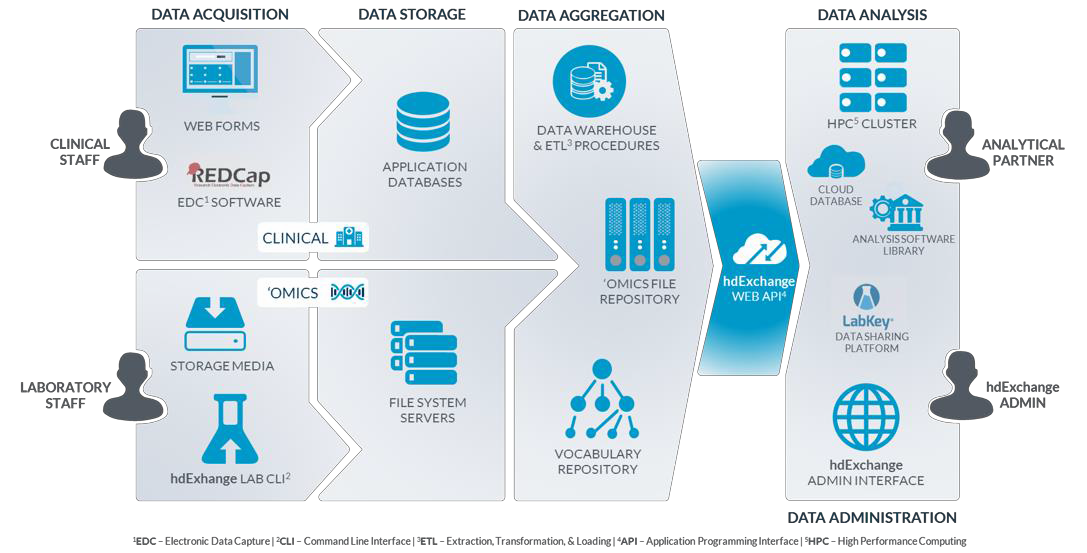
In its role as the TEDDY DCC, the USF Health Informatics Institute (HII) has developed a comprehensive platform for the acquisition, management, integration, analysis, and sharing of scientific data assets, particularly for the Big Data associated with ‘omics research. Access to this data infrastructure for TEDDY and approved external investigators is secured through advanced authentication and an integrated, token-based security model with data encryption. All identifiers are removed or dynamically masked to ensure research integrity and privacy of participants.
The figure below depicts the various components of the data infrastructure developed and employed in support of the TEDDY study. More information can be found in the HII Data Infrastructure slides, hdExchange Platform Overview document, or by emailing TEDDY@epi.usf.edu.
Additional Documentation
Study Overview
Sample and Assay Details
PDF Viewer Required
|



